SMFM Pregnancy Meeting 2020
PUBLISHED ON JAN 22, 2020 — CONFERENCE, POSTER
Hi, thank you for your interest in our prenatal genotyping work. You can find our poster from the SMFM Annual Pregnancy Meeting HERE. Please feel free to contact me regarding the work or potential collaborations. I am also happy to grab a coffee at the meeting if you would like to speak in person.
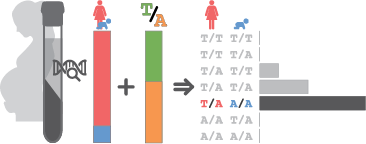
Objective: Congenital abnormalities affect 3-4% of pregnancies and cause 20-30% of neonatal deaths worldwide. Non-invasive prenatal screening (NIPS) leverages the existence of cell-free fetal DNA (cffDNA) with next-generation sequencing to predict fetal presence of large chromosomal abnormalities and, recently, de novo or paternally-inherited dominant SNPs. However, current testing does not give parents information regarding recessive conditions, obscuring over half of known single-gene disorders. Here we present the first sequencing-based approach to detect reliably both dominant and recessive conditions in the fetus using maternal blood.
Study Design: In theory, given the proportion of fetal to maternal DNA (fetal fraction), a true estimate of the allele frequency defines the maternal and fetal genotypes at that locus. We are employing two genotyping panels to interrogate selectively specific genetic variants: (1) an off-the-shelf panel of probes with even density throughout the genome to define the fetal fraction and detect chromosomal abnormalities; (2) a custom panel of probes covering the most common variants in diverse populations. We are currently collecting the first cohort of 100 patients, with the first sequencing results expected in January.1
Results: Using preliminary data we performed 56,000 robust simulations of our genotyping framework. Looking across seven diverse populations, we can estimate the fetal fraction with a novel empirical Bayesian approach with an average estimation error of 0.1%. Using our novel algorithm, we then interrogated genotyping accuracy at maternally heterozygous loci, obtaining >99.5% accuracy for fetal fractions of 5-8% and >99.9% accuracy for fetal fractions >8%.
Conclusions: Using robust simulations we demonstrate the ability to genotype fetuses reliably using only maternal blood. Our expanded NIPS will create a new paradigm for fetal medicine by allowing for precision perinatal treatments based on genomic data.
-
We did not receive the funding we were hoping for until January 31, 2020. As such we do not yet have any sequencing results. We have nearly completed collecting the whole cohort, and plan to have sequencing for the first 50 duos in the near future. ↩︎